MATERIAL AND METHODS
Site Description and Sampling
Sediment cores from Site 1230 (9░6.75┤S, 80░35.0┤W) from the Peru margin were obtained using the advanced piston corer (APC). Site 1230 is located on the lower slope of the Peru Trench in a water depth of 5098 m and contains several horizons of methane hydrate below 70 mbsf. Sediment methane levels were tested on board the ship and ranged from 10 to 1000 ÁM (D'Hondt, J°rgensen, Miller, et al., 2003). Acetate levels at Site 1230 were particularly high at 5–20 ÁM in the sulfate reduction zone (0–8 mbsf) and reached 230 ÁM in the methane zone (8–250 mbsf) at 145 mbsf. Sediment temperature ranged from 2░C at the seafloor and increased to 12░C by 300 mbsf (D'Hondt, J°rgensen, Miller, et al., 2003). Contamination from drilling fluid and seawater in the drill cores was determined to be low or nonexistent (D'Hondt, J°rgensen, Miller, et al., 2003). The uppermost sediment from core 201-1230C-1H, the seafloor sediment, was placed into a mylar bag in the ship's cold room. The bag was flushed with filter-sterilized N2, sealed, and then stored under refrigeration and shipped as described below. Cores of deeper sediment were subsampled as whole round cores, stored under N2 in the dark at 4░C, and shipped cold to The Pennsylvania State University (USA), where they were stored in a cold room at 4░C. Whole-round cores were sampled for microbiological study by removing the outermost 1 cm of sediment and sampling the inner core with sterile syringes under an N2 atmosphere.
Seafloor Enrichments
Marine broth 2216 (Difco) (DMB) medium was prepared according to manufacturer's instructions, and 15 g/L agar was added when plates were used. Aerobic liquid cultures were incubated with shaking at 200 rpm. Seafloor sediment was suspended in liquid media (1 cm3 sediment/5 mL media) on ice, and 100 ÁL of this slurry was immediately plated in duplicate for incubation at both 10░ and 2░C. Selected isolated colonies were restreaked several times to homogeneity. Isolates were maintained on agar at 10░ or 2░C. Genomic DNA was extracted from pure cultures grown in liquid media using the Puregene DNA isolation kit (Gentra Systems) according to the manufacturer's instructions for gram-negative cells. The 16S ribosomal DNA (rDNA) genes were amplified using the general bacterial primer set 8F-1492R and Ready to Go polymerase chain reaction (PCR) beads (Amersham) (Pace et al., 1986; Weisburg, et al., 1991). The amplified products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced at the Pennsylvania State Nucleic Acid Facility on an ABI 3100 sequencer. Sequences are available in the GenBank nucleotide sequence database under accession numbers AY849798–AY849802.
Isolates were examined for their ability to grow at different temperatures on agar media. Optimal growth temperature was estimated by measuring colony size after equivalent incubation times at different temperatures (results from liquid cultures were similar). Growth rates were determined by Klett meter readings of liquid cultures incubated at various temperatures. Four distinct extracellular enzyme activities were assayed, between 2░ and 37░C, as follows: (1) protein hydrolysis was detected by clearing zones on DMB agar with skim milk added (15 g/L) (Lichenstein et al., 1992); (2) esterase activity was detected by precipitation zones on DMB agar supplemented with 1% Tween-20 (ICN Biomedicals) (Jensen, 1983); (3) chitin hydrolysis was detected by clearing zones on DMB agar containing colloidal chitin (15g/L) (Lingappa and Lockwood, 1961; Morgavi et al., 1994); and (4) starch hydrolysis was indicated by clear zones on iodine-stained DMB agar with starch (15g/L).
Enrichment of Mixed Populations
Sediment from 0.67 mbsf (Section 201-1230C-1H-1) was inoculated (2 cm3/50 mL) into aerobic liquid DMB and incubated at 10░C. Mixed populations were then transferred to both aerobic DMB and anaerobic DMB (N2 atmosphere). After incubating for 3 weeks, the mixed population was passed through the following enrichment and dilution steps to encourage growth of possible facultative anaerobes. Each step included inoculation of 1 mL culture into 50 mL of media and alternated between aerobic and anaerobic media. All enrichments were incubated for 1 month at 2░C prior to further transfer. At the sixth month, the enrichment culture was diluted 1:108 into anaerobic DMB and sampled after 1 month in an attempt to achieve a pure culture. Microscopic observations were made using wet mounts and viewed at 1000x under oil immersion on a MicroMaster phase contrast microscope.
DNA was extracted from the mixed populations using the UltraClean Microbial DNA isolation kit (MoBio) according to manufacturer's instructions. Ribosomal intergenic spacer analysis (RISA) was performed with bacterial primers 16S 1406F–23S 115R or archaeal primers 16S 1214F–23S 46R and amplified by Ready-to-Go beads (Amersham) (annealing temperature = 59░C) (Lane, 1991). The RISA products were analyzed on 3% TAE-agarose gels. The 16S rDNA genes were amplified from total extracted DNA purified as described above. The 16S rDNA libraries were prepared using the PCR-script Amp cloning kit (Stratagene) and were transformed into Z-competent Escherichia coli DH5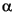 cells (Zymo, Inc.). Transformants containing inserts were identified by blue/white screening and the plasmids extracted using the Wizard Plus SV Miniprep kit (Promega). Plasmid inserts were screened using amplified ribosomal DNA restriction analysis (ARDRA) and grouped by digestion pattern by RsaI (Promega). At least one representative of each pattern was sequenced as described above. Comparable sequences from GenBank (www.ncbi.nlm.nih.gov) and RDPII (rdp.cme.msu.edu) (Cole et al., 2003) databases were gathered. Phylogenetic trees were prepared by aligning these by Clustal X found in the BioEdit platform (www.mbio.ncsu.edu/BioEdit/bioedit.html) and tree preparation by PAUP (version 4.0b10; paup.csit.fsu.edu). Bootstrap values represent 100 replicates.
cells (Zymo, Inc.). Transformants containing inserts were identified by blue/white screening and the plasmids extracted using the Wizard Plus SV Miniprep kit (Promega). Plasmid inserts were screened using amplified ribosomal DNA restriction analysis (ARDRA) and grouped by digestion pattern by RsaI (Promega). At least one representative of each pattern was sequenced as described above. Comparable sequences from GenBank (www.ncbi.nlm.nih.gov) and RDPII (rdp.cme.msu.edu) (Cole et al., 2003) databases were gathered. Phylogenetic trees were prepared by aligning these by Clustal X found in the BioEdit platform (www.mbio.ncsu.edu/BioEdit/bioedit.html) and tree preparation by PAUP (version 4.0b10; paup.csit.fsu.edu). Bootstrap values represent 100 replicates.
A basal anaerobic marine salts media, DGH (D'Hondt, J°rgensen, Miller et al., 2003), modified from Desulfotomaculum geothermicum media (Daumas et al., 1988), was prepared with the addition of one of four carbon sources: 5 g/L trimethylamine, 8 ml/L methanol, 8 g/L acetate, or 2-atm CO2. Media were reduced with sodium sulfide, degassed under N2/CO2, and aliquoted under an N2/H2 glovebag atmosphere; the headspace was then exchanged for either 1.5-atm N2/CO2 or 2-atm H2/CO2. Resazurin was used as an oxygen indicator. Sediment was added to make an anaerobic slurry preparation of 1 cm3 of sediment in 9 mL DGH media with no carbon source; 1 mL of this was added to 10 mL media for incubation. Enrichment cultures were incubated at 4░C without shaking. Growth was monitored by analysis of headspace gas on a HP 5890 Series II gas chromatograph. DNA analysis was performed as described above for mixed populations.

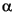 cells (Zymo, Inc.). Transformants containing inserts were identified by blue/white screening and the plasmids extracted using the Wizard Plus SV Miniprep kit (Promega). Plasmid inserts were screened using amplified ribosomal DNA restriction analysis (ARDRA) and grouped by digestion pattern by RsaI (Promega). At least one representative of each pattern was sequenced as described above. Comparable sequences from GenBank (www.ncbi.nlm.nih.gov) and RDPII (rdp.cme.msu.edu) (Cole et al., 2003) databases were gathered. Phylogenetic trees were prepared by aligning these by Clustal X found in the BioEdit platform (www.mbio.ncsu.edu/BioEdit/bioedit.html) and tree preparation by PAUP (version 4.0b10; paup.csit.fsu.edu). Bootstrap values represent 100 replicates.
cells (Zymo, Inc.). Transformants containing inserts were identified by blue/white screening and the plasmids extracted using the Wizard Plus SV Miniprep kit (Promega). Plasmid inserts were screened using amplified ribosomal DNA restriction analysis (ARDRA) and grouped by digestion pattern by RsaI (Promega). At least one representative of each pattern was sequenced as described above. Comparable sequences from GenBank (www.ncbi.nlm.nih.gov) and RDPII (rdp.cme.msu.edu) (Cole et al., 2003) databases were gathered. Phylogenetic trees were prepared by aligning these by Clustal X found in the BioEdit platform (www.mbio.ncsu.edu/BioEdit/bioedit.html) and tree preparation by PAUP (version 4.0b10; paup.csit.fsu.edu). Bootstrap values represent 100 replicates.