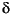 14C analysis. Geochim. Cosmochim. Acta, 65:3123–3137.
doi:10.1016/S0016-7037(01)00657-3
14C analysis. Geochim. Cosmochim. Acta, 65:3123–3137.
doi:10.1016/S0016-7037(01)00657-3
Amann, R.I., Ludvig, W., and Schleifer, K.H., 1995. Physiological identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev., 59:143–169.
Bale, S.J., Goodman, K., Rochelle, P.A., Marchesi, J.R., Fry, J.C., Weightman, A.J., and Parkes, R.J., 1997. Desulfovibrio profundus sp. nov., a novel barophilic sulfate-reducing bacterium from deep sediment layers in the Japan Sea. Int. J. Syst. Bacteriol., 47:515–521.
Barnes, S.P., Bradbrook, S.D., Cragg, B.A., Marchesi, J.R., Weightman, A.J., Fry, J.C., and Parkes, R.J., 1998. Isolation of sulfate-reducing bacteria from deep sediment layers of the Pacific Ocean. Geomicrobiol. J., 15:67–83.
Barns, S.M., Delwiche, C.F., Palmer, J.D., and Pace, N.R., 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA, 93:9188–9193.
Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Rapp, B.A., and Wheeler, D.L., 2000. GenBank. Nucleic Acids Res., 28:15–18.
Biddle, J.F., Lipp, J.S., Lever, M., Lloyd, K., Sørensen, K., Anderson, R., Fredricks, H.F., Elvert, M., Kelly, T.J., Schrag, D.P., Sogin, M.L., Brenchley, J.E., Teske, A., House, C.H., and Hinrichs, K.-U., 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. USA, 103(10):3846–3851. doi:10.1073/pnas.0600035103
Boetius, A., Ravenschlag, K., Schubert, C.J., Rickert, D., Widdel, F., Gieseke, A., Amann, R., Jørgensen, B.B., Witte, U., and Pfannkuche, O., 2000. A marine microbial consortium apparently mediating the anaerobic oxidation of methane. Nature (London, U. K.), 407:623–626. doi:10.1038/35036572
Boone, D.R., and Mah, R.A., 2001. Genus I. Methanosarcina. In Garrity, G.M., and Boone, D.R. (Eds.), Bergey's Manual of Systematic Bacteriology (2nd ed.). Vol. 1. The Archaea and the Deeply Branching and Phototrophic Bacteria: New York (Springer), 269–276.
Braun, M., Mayer, F., and Gottschalk, G., 1981. Clostridium aceticum (Wieringa), a microorganism producing acetic acid from molecular hydrogen and carbon dioxide. Arch. Microbiol., 128:288–293.
Cano, R.J., and Borucki, M.K,. 1995. Revival and identification of bacterial spores in 25–40 million-year-old Dominican amber. Science, 268:1060–1064.
Chandler, D.P., Brockman, F.J., Bailey, T.J., and Fredrickson, J.K., 1998. Phylogenetic diversity of archaea and bacteria in a deep subsurface paleosol. Microb. Ecol., 36:37–50. doi:10.1007/s002489900091
Coolen, M.J.L., Cypionka, H., Sass, A.M., Sass, H., and Overmann, J., 2002, Ongoing modification of Mediterranean Pleistocene sapropels mediated by prokaryotes. Science, 296:2407–2410. doi:10.1126/science.1071893
DeLong, E.F., 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U. S. A., 89:5685–5689.
DeLong, E.F., 1998. Everything in moderation: archaea as "non-extremophiles." Curr. Opin. Genet. Dev., 8:649–654. doi:10.1016/S0959-437X(98)80032-4
DeLong, E.F., Wu, K.Y., Prezelin, B.B., and Jovine, R.V.M., 1994. High abundance of archaea in Antarctic marine picoplankton. Nature (London, U. K.), 371:695–697. doi:10.1038/371695a0
D'Hondt, S.D., Jørgensen, B.B., Miller, D.J., Batzke, A., Blake, R., Cragg, B.A., Cypionka, H., Dickens, G.R., Ferdelman, T., Hinrichs, K.-H., Holm, N.G., Mitterer, R., Spivack, A., Wang, G., Bekins, B., Engelen, B., Ford, K., Gettemy, G., Rutherford, S.D., Sass, H., Skilbeck, C.G., Aiello, I.W., Guerin, G., House, C.H., Inagak, F., Meister, P., Naehr, T., Niitsuma, S., Parkes, R.J., Schippers, A., Smith, D.C., Teske, A., Wiegel, J., Naranjo Padillo, C., and Solis Acosta, J.L., 2004. Distributions of microbial activities in deep subseafloor sediments. Science, 306:2216–2221. doi:10.1126/science.1101155
D'Hondt, S.L., Jørgensen, B.B., Miller, D.J., et al., 2003. Proc. ODP, Init. Repts., 201 [CD-ROM]. Available from: Ocean Drilling Program, Texas A&M University, College Station TX 77845-9547, USA. [HTML]
D'Hondt, S.D., Rutherford, S., and Spivack, A.J., 2002. Metabolic activity of the subsurface biosphere in deep-sea sediments. Science, 295:2067–2070. doi:10.1126/science.1064878
Fuhrman, J.A., McCallum, K., and Davis, A.A., 1992. Novel archaebacterial group from marine plankton. Nature (London, U. K.), 356:148–149. doi:10.1038/356148a0
Fuhrman, J.A., and Ouverney, C.C., 1998. Marine microbial diversity studied via 16S RNA sequences: cloning results from coastal waters and counting of native archaea with fluorescent single cell probes. Aquat. Ecol., 32:3–15. doi:10.1023/A:1009974817127
Fukui, M., Suwa, Y., and Urushigawa, Y., 1996. High survival efficiency and ribosomal RNA decaying pattern of Desulfobacter latus, a highly specific acetate-utilizing organism, during starvation. FEMS Microbiol. Ecol., 19:17–25. doi:10.1111/j.1574-6941.1996.tb00194.x
Gest, H., and Mandelstam, J., 1987. Longevity of microorganisms in natural environments. Microbiol. Sci., 4:69–71.
Hales, B.A., Edwards, C., Ritchie, D.A., Hall, G., Pickup, R.W., and Saunders, J.R., 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol., 62:668–675.
Hallam, S.J., Girguis, P.R., Preston, C.M., Richardson, P.M., and DeLong, E.F., 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol., 69:5483–5491. doi:10.1128/AEM.69.9.5483-5491.2003
Hinrichs, K.-U., Hayes, J.M., Sylva, S.P., Brewer, P.G., and DeLong, E.F., 1999. Methane consuming archaebacteria in marine sediments. Nature (London, U. K.), 398:802–805. doi:10.1038/19751
Huber, J.A., Butterfield, D.A., and Baross, J.A., 2002. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol., 68:1585–1594. doi:10.1128/AEM.68.4.1585-1594.2002
Hugenholtz, P., 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol., 3(2). doi:10.1186/gb-2002-3-2-reviews0003
Hugenholtz, P., Goebel, B.M., and Pace, N.R., 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol., 180:4765–4774.
Inagaki, F., Nunoura, T., Nakagawa, S., Teske, A., Lever, M., Lauer, A., Suzuki, M., Takai, K., Delwiche, M., Colwell, F.S., Nealson, K.H., Horikoshi, K., D'Hondt, S.L., and Jørgensen, B.B., 2006. Biogeographical distribution and diversity of microbes in methane hydrate–bearing deep marine sediments on the Pacific Ocean margin. Proc. Natl. Acad. Sci. USA, 103:2815–2820. doi:10.1073/pnas.0511033103
Inagaki, F., Okada, H., Tsapin, A.I., and Nealson, K.H., 2005. Microbial survival: the paleome: a sedimentary genetic record of past microbial communities. Astrobiology, 5(2):141–153. doi:10.1089/ast.2005.5.141
Inagaki, F., Suzuki, M., Takai, K., Oida, H., Sakamoto, T., Aoki, K., Nealson, K.H., and Horikoshi, K., 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol., 69:7224–7235. doi:10.1128/AEM.69.12.7224-7235.2003
Isaksen, M.F., Bak, F., and Jørgensen, B.B., 1994. Thermophilic sulfate-reducing bacteria in cold marine sediment. FEMS Microbiol. Ecol., 14:1–8. doi:10.1111/j.1574-6941.1994.tb00084.x
Karner, M.B., DeLong, E.F., and Karl, D.M., 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature (London, U. K.), 409:507–510. doi:10.1038/35054051
Klein, M., Friedrich, M., Roger, A.J., Hugenholtz, P., Fishbain, S., Abicht, H., Blackall, L.L., Stahl, D.A., and Wagner, M., 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol., 183:6028–6035. doi:10.1128/JB.183.20.6028-6035.2001
Könneke, M., Bernhard, A.E., de la Torre, J.R., Walker, C.B., Waterbury, J.B., and Stahl, D.A., 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature (London, U. K.), 437:543–546. doi:10.1038/nature03911
Kormas, K.A., Smith, D.C., Edgcomb, V., and Teske, A., 2003. Molecular analysis of deep subsurface microbial communities in Nankai Trough sediments (ODP Leg 190, Site 1176). FEMS Microbiol. Ecol., 45(2):115–125. doi:10.1016/S0168-6496(03)00128-4
Lee, Y.-J., Wagner, I.D., Brice, M.E., Kevbrin, V.V., Mills, G.L., Romanek, C.S., and Wiegel, J., 2005. Thermosediminibacter oceani gen. nov., sp. nov. and Thermosediminibacter litoriperuensis sp. nov., new anaerobic thermophilic bacteria isolated from Peru margin. Extremophiles, 9:375–383. doi:10.1007/s00792-005-0453-4
Lever, M., and Teske, A., 2005. Deep marine subsurface microbial communities of the Peru Trench: sulfate reducers, methanogens, and unknowns. Astrobiology, 5:259. (Abstract)
Luton, P.E., Wayne, J.M., Sharp, R.J., and Riley, P.W., 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology, 148:3521–3530.
Marchant, R., Banat, I.M., Rahman, T.J., and Berzano, M., 2002. The frequency and characteristics of highly thermophilic bacteria in cool soil environments. Environ. Microbiol., 4:595–602. doi:10.1046/j.1462-2920.2002.00344.x
Marchesi, J.R., Weightman, A.J., Cragg, B.A., Parkes, R.J., and Fry, J.C., 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol., 34:221–228. doi:10.1111/j.1574-6941.2001.tb00773.x
Mauclaire, L., Zepp, K., Meister, P., and McKenzie, J., 2005. Direct in situ detection of cells in deep-sea sediment cores from the Peru margin (ODP Leg 201, Site 1229). Geobiology, 2:217–223. doi:10.1111/j.1472-4677.2004.00035.x
Maymó-Gatell, X., Chien, Y.-T., Gossett, J.M., and Zinder, S.H., 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethane. Science, 276:1568–1571. doi:10.1126/science.276.5318.1568
Michaelis, W., Seifert, R., Nauhaus, K., Treude, T., Thiel, V., Blumenberg, M., Knittel, K., Gieseke, A., Peterknecht, K., Pape, T., Boetius, A., Amann, R., Jørgensen, B.B., Widdel, F., Peckmann, J., Pimenov, N.V., and Gulin, M.B., 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science, 297:1013–1015. doi:10.1126/science.1072502
Miller, T.L., 2001. Genus II. Methanobrevibacter. In Garrity, G.M., and Boone, D.R. (Eds.), Bergey's Manual of Systematic Bacteriology (2nd ed.). Vol. 1. The archaea and the deeply branching and phototrophic bacteria. New York (Springer), 219–226.
Molin, S., and Givskov, M., 1999. Application of molecular tools for in-situ monitoring of bacterial growth activity. Environ. Microbiol., 1:383–391. doi:10.1046/j.1462-2920.1999.00056.x
Newberry, C.J., Webster, G., Cragg, B.A., Parkes, R.J., Weightman, A.J., and Fry, J.C., 2004. Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ. Microbiol., 6:274–287. doi:10.1111/j.1462-2920.2004.00568.x
Orphan, V., House, C.H., Hinrichs, K.-U., McKeegan, K.D., and DeLong, E.F., 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA, 99:7663–7668.
Orphan, V.J., House, C.H., Hinrichs, K.-U., McKeegan, K.D., and DeLong, E.F., 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science, 293:484–487. doi:10.1126/science.1061338
Ouverney, C.C., and Fuhrman, J.A., 2000. Marine planktonic archaea take up amino acids. Appl. Environ. Microbiol., 66:4829–4833. doi:10.1128/AEM.66.11.4829-4833.2000
Parkes, R., Cragg, B., and Wellsbury, P., 2000. Recent studies on bacterial populations and processes in subseafloor sediments: a review. Hydrogeol. J., 8:11–28.
Parkes, R.J., Webster, G., Cragg, B.A., Weightman, A.J., Newberry, C.J., Ferdelman, T.G., Kallmeyer, J., Jørgensen, B.B., Aiello, I.W., and Fry, J.C., 2005. Deep sub-seafloor prokaryotes stimulated at interfaces over geological time. Nature (London, U. K.), 436:390–394. doi:10.1038/nature03796
Patel, G.B., 2001. Genus I. Methanosaeta. In Garrity, G.M., and Boone, D.R. (Eds.), Bergey's Manual of Systematic Bacteriology (2nd ed.). Vol. 1. The Archaea and the deeply branching and phototrophic bacteria. New York (Springer), 289–294.
Pearson, A., McNichol, A.P., Benitez-Nelson, B.C., Hayes, J.M., and Eglinton, T.I., 2001. Origins of lipid biomarkers in Santa Monica Basin surface sediment: a case study using compound-specific 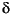 14C analysis. Geochim. Cosmochim. Acta, 65:3123–3137.
doi:10.1016/S0016-7037(01)00657-3
14C analysis. Geochim. Cosmochim. Acta, 65:3123–3137.
doi:10.1016/S0016-7037(01)00657-3
Pernthaler, J., and Amann, R., 2005. Fate of heterotrophic microbes in pelagic habitats: focus on populations. Microbiol. Mol. Biol. Rev., 69:440–461. doi:10.1128/MMBR.69.3.440-461.2005
Perhthaler, A., Pernthaler, J., and Amann, R., 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol., 68(6):3094–3101. doi:10.1128/AEM.68.6.3094-3101.2002
Poulsen, L.K., Ballard, G., and Stahl, D.A., 1993. Use of ribosomal-RNA fluorescence in-situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol., 59:1354–1360.
Rappe, M.S., and Giovannoni, S.J., 2003. The uncultured microbial majority. Annu. Rev. Microbiol., 57:369–394.
Reed, D.W., Fujita, Y., Delwiche, M.E., Blackwelder, D.B., Sheridan, P.P., Uchida, T., and Colwell, F.S., 2002. Microbial communities from methane hydrate–bearing deep marine sediments in a forearc basin. Appl. Environ. Microbiol., 68:3759–3770. doi:10.1128/AEM.68.8.3759-3770.2002
Schippers, A., Neretin, L.N., Kallmeyer, J., Ferdelman, T.G., Cragg, B.A., Parkes, J.R., and Jørgensen, B.B., 2005. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature (London, U. K.), 433:861–864. doi:10.1038/nature03302
Schrenk, M.O., Kelley, D.S., Delaney, J.R., and Baross, J.A., 2003. Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl. Environ. Microbiol., 69:3580–3592. doi:10.1128/AEM.69.6.3580-3592.2003
Sekiguchi, Y., Takahashi, H., Kamagata, Y., Ohashi, A., and Harada, H., 2001. In situ detection, isolation, and physiological properties of a thin filamentous microorganism abundant in methanogenic granular sludges: a novel isolate affiliated with a clone cluster, the green non-sulfur bacteria, subdivision I. Appl. Environ. Microbiol., 67:5740–5749. doi:10.1128/AEM.67.12.5740-5749.2001
Sekiguchi, Y., Yamada, T., Hanada, S., Ohashi, A., Harada, H., and Kamagata, Y., 2003. Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol., 53:1843–1851. doi:10.1099/ijs.0.02699-0
Sørensen, K.B., Lauer, A., and Teske, A., 2004. Archaeal phylotypes in a metal-rich, low-activity deep subsurface sediment of the Peru Basin, ODP Leg 201, Site 1231. Geobiology, 2:151–161. doi:10.1111/j.1472-4677.2004.00028.x
Sørensen, K.B., and Teske, A., in press. Stratified communities of active archaea in geochemically distinct deep marine subsurface sediments. Appl. Environ. Microbiol.
Springer, E., Sachs, M.S., Woese, C.R., and Boone, D.R., 1995. Partial gene sequences for the alpha-subunit of methyl-coenzyme M reductase (MCR1) as a phylogenetic tool for the family Methanosarcinaceae. Int. J. Syst. Bacteriol., 45:554–559.
Stein, L.Y., Jones, G., Alexander, B., Elmund, K., Wright-Jones, C., and Nealson, K.H., 2002. Intriguing microbial diversity associated with metal-rich particles from a freshwater reservoir. FEMS Microbiol. Ecol., 42:431–440. doi:10.1111/j.1574-6941.2002.tb01032.x
Sturt, H.F., Summons, R.E., Smith, K., Elvert, M., and Hinrichs, K.-U., 2004. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom., 18:617–628. doi:10.1002/rcm.1378
Suess, J., Engelen, B., Cypionka, H., and Sass, H., 2004. Quantitative analysis of bacterial communities from Mediterranean sapropels based on cultivation-dependent methods. FEMS Microbiol. Ecol., 51:109–121. doi:10.1016/j.femsec.2004.07.010
Suess, J., Sass, H., Cypionka, H., and Engelen, B., 2005. Widespread distribution of Rhizobium radiobacter in Mediterranean sediments. Progr. Abstr., Jnt. Int. Symp. Subsurf. Microbiol. (ISSM 2005) and Environ. Biogeochem. (ISEB XVII), 63.
Takai, K., and Horikoshi, K., 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics, 152:1285–1297.
Takai, K., Moser, D.P., DeFlaun, M., Onstott, T.C., and Fredrickson, J.K., 2001. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol., 67:5750–5760. doi:10.1128/AEM.67.21.5750-5760.2001
Teira, E., Reinthaler, T., Pernthaler, A., Pernthaler, J., and Herndl, G.J., 2004. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl. Environ. Microbiol., 70(7):4411–4414. doi:10.1128/AEM.70.7.4411-4414.2004
Teske, A., Dhillon, A., and Sogin, M.S., 2003. Genomic markers of ancient anaerobic microbial pathways: sulfate reduction, methanogenesis, and methane oxidation. Biol. Bull. (Woods Hole, MA, U. S.), 204:186–191.
Teske, A., Hinrichs, K.-U., Edgcomb, V., Gomez, A.d.V., Kysela, D., Sylva, S.P., Sogin, M.L., and Jannasch, H.W., 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol., 68:1994–2007. doi:10.1128/AEM.68.4.1994-2007.2002
Vetriani, C., Jannash, H.W., MacGregor, B.J., Stahl, D.A., and Reysenbach, A.L., 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol., 65:4375–4384.
Wagner, M., Roger, A.J., Flax, J.L., Brusseau, G.A., and Stahl, D.A., 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol., 180:2975–2982.
Webster, G., Newberry, C.J., Fry, J.C., and Weightman, A.J., 2003. Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: a cautionary tale. J. Microbiol. Methods, 55:155–164. doi:10.1016/S0167-7012(03)00140-4
Webster, G., Parkes, R.J., Fry, J.C., and Weightman, A.J., 2004. Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl. Environ. Microbiol., 70(9):5708–5713. doi:10.1128/AEM.70.9.5708-5713.2004
Woese, C.R., 1987. Bacterial evolution. Microbiol. Rev., 51:221–271.
Woese, C.R., Kandler, O., and Wheelis, M., 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA, 87:4576–4579.
Whitman, W.B., Coleman, D.C., and Wiebe, W.J., 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA, 95:6578–6583. doi:10.1073/pnas.95.12.6578
Wuchter, C., Schouten, S., Boschker, H.T.S., and Damsté, J.S.S., 2003. Bicarbonate uptake by marine crenarchaeota. FEMS Microbiol. Lett., 219:203–207. doi:10.1016/S0378-1097(03)00060-0
Zink, K.-G., Wilkes, H., Disko, U., Elvert, M., and Horsfield, B., 2003. Intact phospholipids–microbial "life markers" in marine deep subsurface sediments. Org. Geochem., 34:755–769. doi:10.1016/S0146-6380(03)00041-X
Zverlov, V., Klein, M., Lucker, S., Friedrich, M.W., Kellermann, J., Stahl, D.A., Loy, A., and Wagner, M., 2005. Lateral gene transfer of dissimilatory (bi)sulfite reductase revisited. J. Bacteriol., 187:2203–2208. doi:10.1128/JB.187.6.2203-2208.2005